t-distributed Stochastic Neighbor Embedding (t-SNE)
Explore how t-SNE visualizes high-dimensional data. Learn to reveal clusters in computer vision features for Ultralytics YOLO26 and optimize machine learning models.
t-distributed Stochastic Neighbor Embedding (t-SNE) is a statistical method for visualizing high-dimensional data by
giving each data point a location in a two or three-dimensional map. This technique, a form of non-linear
dimensionality reduction, is widely used
in machine learning to explore datasets that contain hundreds or thousands of features. Unlike linear methods that
focus on preserving global structures, t-SNE excels at keeping similar instances close together, revealing local
clusters and manifolds that might otherwise remain hidden. This makes it an invaluable tool for everything from
genomic research to understanding the internal logic of deep neural networks.
How t-SNE Works
The core idea behind t-SNE involves converting the similarities between data points into joint probabilities. In the
original high-dimensional space, the algorithm measures the similarity between points using a Gaussian distribution.
If two points are close together, they have a high probability of being "neighbors." The algorithm then
attempts to map these points to a lower-dimensional space (usually 2D or 3D) while maintaining these probabilities.
To achieve this, it defines a similar probability distribution in the lower-dimensional map using a Student's
t-distribution. This specific distribution has heavier tails than a normal Gaussian distribution, which helps address
the "crowding problem"—a phenomenon where points in high-dimensional space tend to collapse on top of each
other when projected down. By pushing dissimilar points farther apart in the visualization, t-SNE creates distinct,
readable clusters that reveal the underlying
structure of the training data. The algorithm
effectively learns the best map representation through
unsupervised learning by minimizing the
divergence between the high-dimensional and low-dimensional probability distributions.
Real-World Applications in AI
t-SNE is a standard tool for
exploratory data analysis (EDA) and model
diagnostics. It allows engineers to "see" what a model is learning.
-
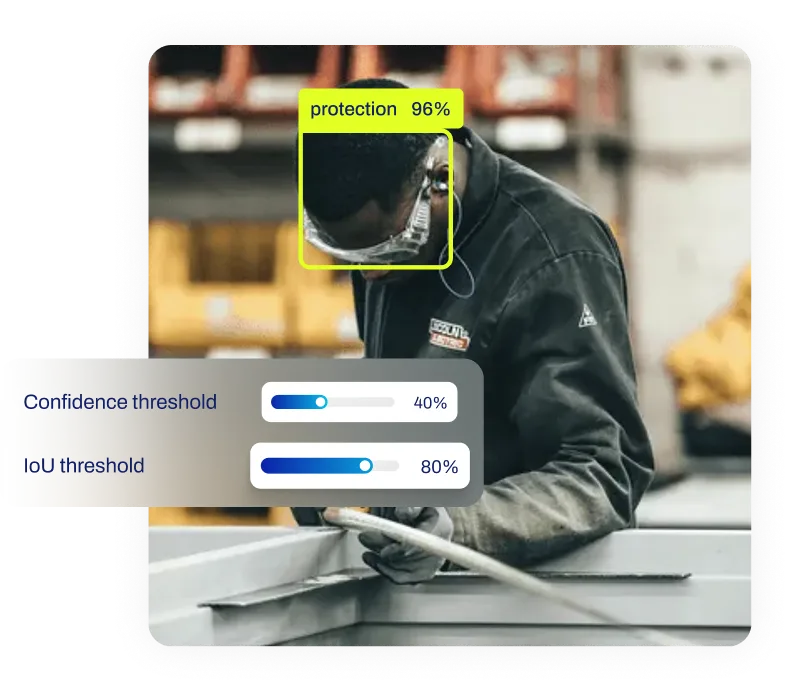
Verifying Computer Vision Features: In object detection workflows using models like
YOLO26, developers often need to check if the network can
distinguish between visually similar classes. By extracting the
feature maps from the final layers of the network
and projecting them with t-SNE, engineers can visualize whether images of "cats" cluster separately from
"dogs." If the clusters are mixed, it suggests the model's
feature extraction capabilities need
improvement.
-
Natural Language Processing (NLP): t-SNE is heavily utilized for visualizing word
embeddings. When high-dimensional word vectors (often
300+ dimensions) are projected into 2D, words with similar semantic meanings naturally group together. For instance,
a t-SNE plot might show a cluster containing "king," "queen," "prince," and
"monarch," demonstrating that the
Natural Language Processing (NLP)
model grasps the concept of royalty.
-
Genomics and Bioinformatics: Researchers use t-SNE to visualize single-cell RNA sequencing data. By
reducing thousands of gene expression values into a 2D plot, scientists can identify distinct cell types and trace
developmental trajectories, aiding in the discovery of new biological insights and disease markers.
Comparison with PCA
It is important to distinguish t-SNE from
Principal Component Analysis (PCA), another common reduction technique.
-
PCA is a linear technique that focuses on preserving the global variance of the data. It is
deterministic and computationally efficient, making it excellent for initial data compression or noise reduction.
-
t-SNE is a non-linear technique focused on preserving local neighborhoods. It is probabilistic
(stochastic) and computationally heavier, but it produces far better visualizations for complex, non-linear
manifolds.
A common best practice in data preprocessing is
to use PCA first to reduce the data to a manageable size (e.g., 50 dimensions) and then apply t-SNE for the final
visualization. This hybrid approach reduces computational load and filters out noise that might degrade the t-SNE
result.
Python Example: Visualizing Features
The following example demonstrates how to use scikit-learn to apply t-SNE to a synthetic dataset. This
workflow mirrors how one might visualize features extracted from a deep learning model.
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from sklearn.manifold import TSNE
# Generate synthetic high-dimensional data (100 samples, 50 features, 3 centers)
X, y = make_blobs(n_samples=100, n_features=50, centers=3, random_state=42)
# Apply t-SNE to reduce dimensions from 50 to 2
# 'perplexity' balances local vs global aspects of the data
tsne = TSNE(n_components=2, perplexity=30, random_state=42)
X_embedded = tsne.fit_transform(X)
# Plot the result to visualize the 3 distinct clusters
plt.scatter(X_embedded[:, 0], X_embedded[:, 1], c=y)
plt.title("t-SNE Projection of High-Dimensional Data")
plt.show()
Key Considerations
While powerful, t-SNE requires careful
hyperparameter tuning. The
"perplexity" parameter is critical; it essentially guesses how many close neighbors each point has. Setting
it too low or too high can result in misleading visualizations. Furthermore, t-SNE does not preserve global distances
well—meaning the distance between two distinct clusters on the plot does not necessarily reflect their physical
distance in the original space. Despite these nuances, it remains a cornerstone technique for validating
computer vision (CV) architectures and
understanding complex datasets. Users managing large-scale datasets often leverage the
Ultralytics Platform to organize their data before performing such
in-depth analysis.









.webp)